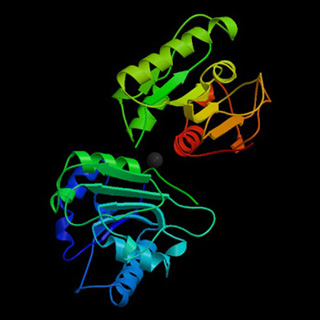
The Crystal Structure Of Bacillus Subtilis Lipase: A Minimal a/b Hydrolase Enzyme. PDB ID: 1I6W. van Pouderoyen, G., T. Eggert, K. E. Jaeger, B. W. Dijkstra: The crystal structure of Bacillus subtilis lipase: a minimal alpha/beta hydrolase fold enzyme. J. Mol. Biol. 309, issue 1 (May 25, 2001): 215-26. (Image courtesy of the Research Collaboratory for Structural Bioinformatics. Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. "The Protein Data Bank." Nucleic Acids Research 28 (2000): 235-242.)
Instructor(s)
Prof. Kristala L. Jones Prather
MIT Course Number
10.492-2
As Taught In
Fall 2004
Level
Undergraduate
Course Description
Course Features
Course Description
This course provides a brief introduction to the field of biocatalysis in the context of process design. Fundamental topics include why and when one may choose to use biological systems for chemical conversion, considerations for using free enzymes versus whole cells, and issues related to design and development of bioconversion processes. Biological and engineering problems are discussed as well as how one may arrive at both biological and engineering solutions.
Other Versions
Other OCW Versions
OCW has published multiple versions of this subject. ![]()


